Answers that enhance clinical insight
Clear results with robust supporting data
Clinicians receive clonoSEQ test results via an informative clinical report. These visually engaging reports provide clinicians with a clear MRD test result while also incorporating rich, quantitative data and comprehensive analysis for clinicians who want to dig deeper.
Report content tailored to inform decisions
Explore example clonoSEQ reports:
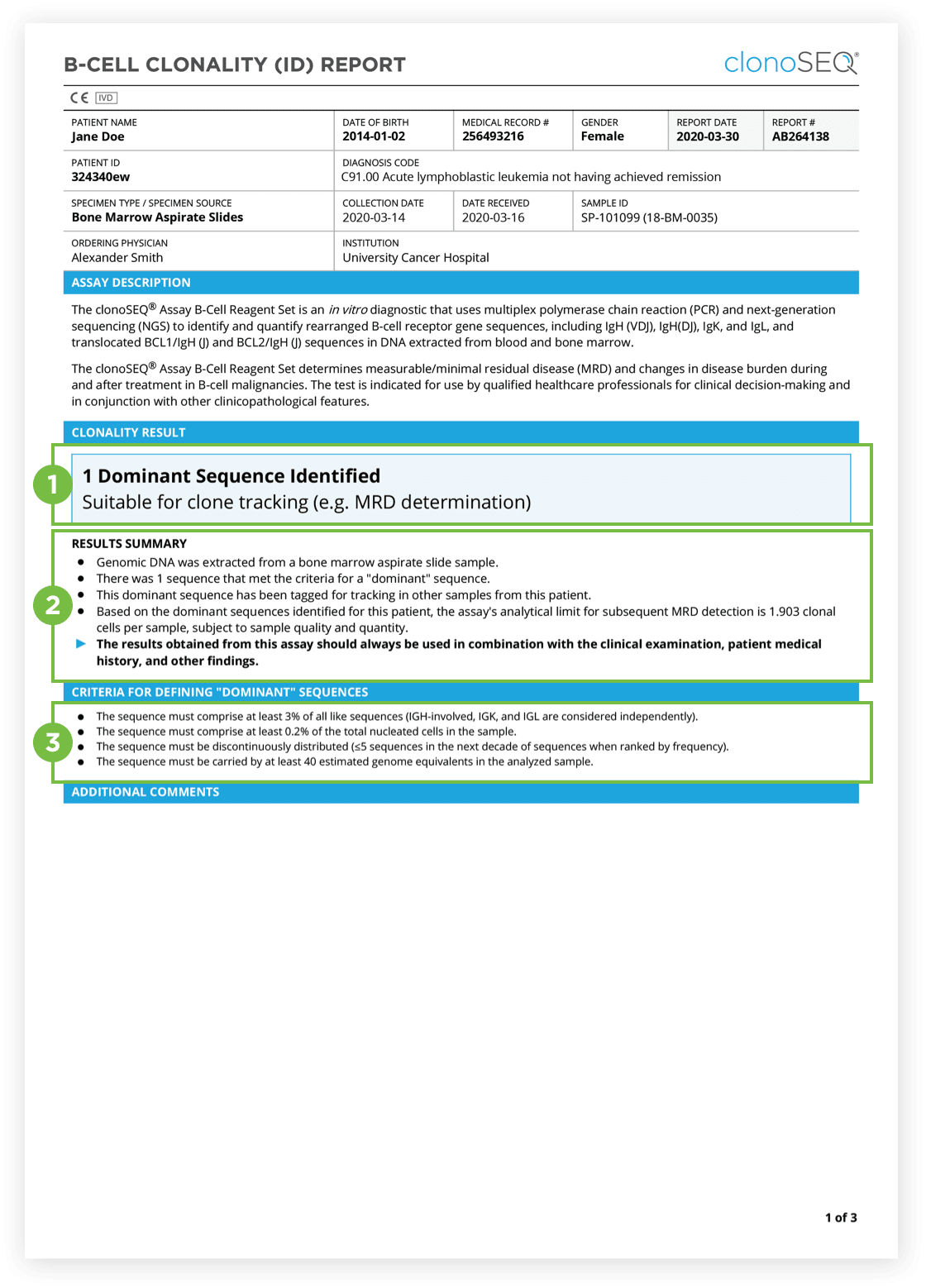
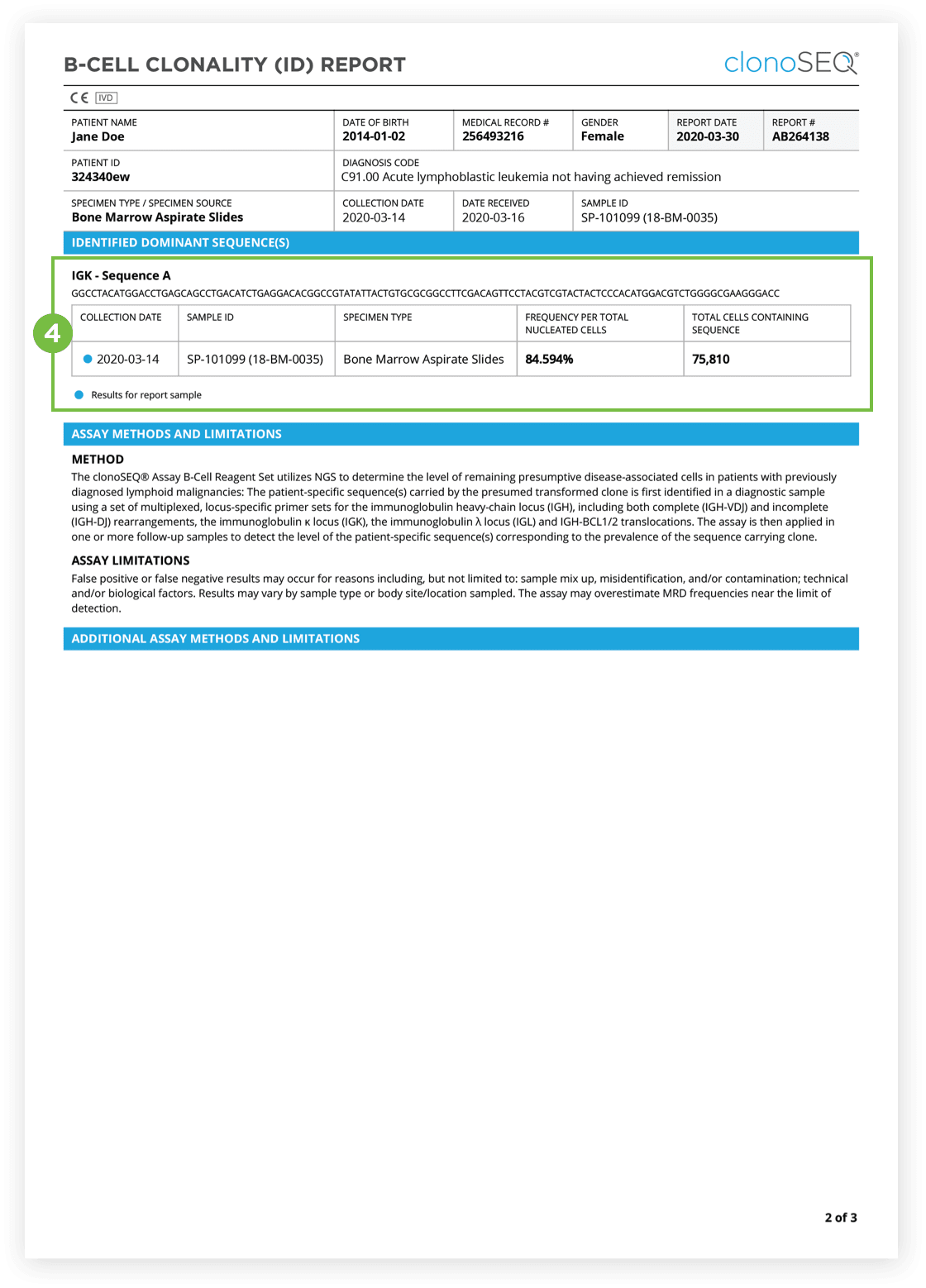
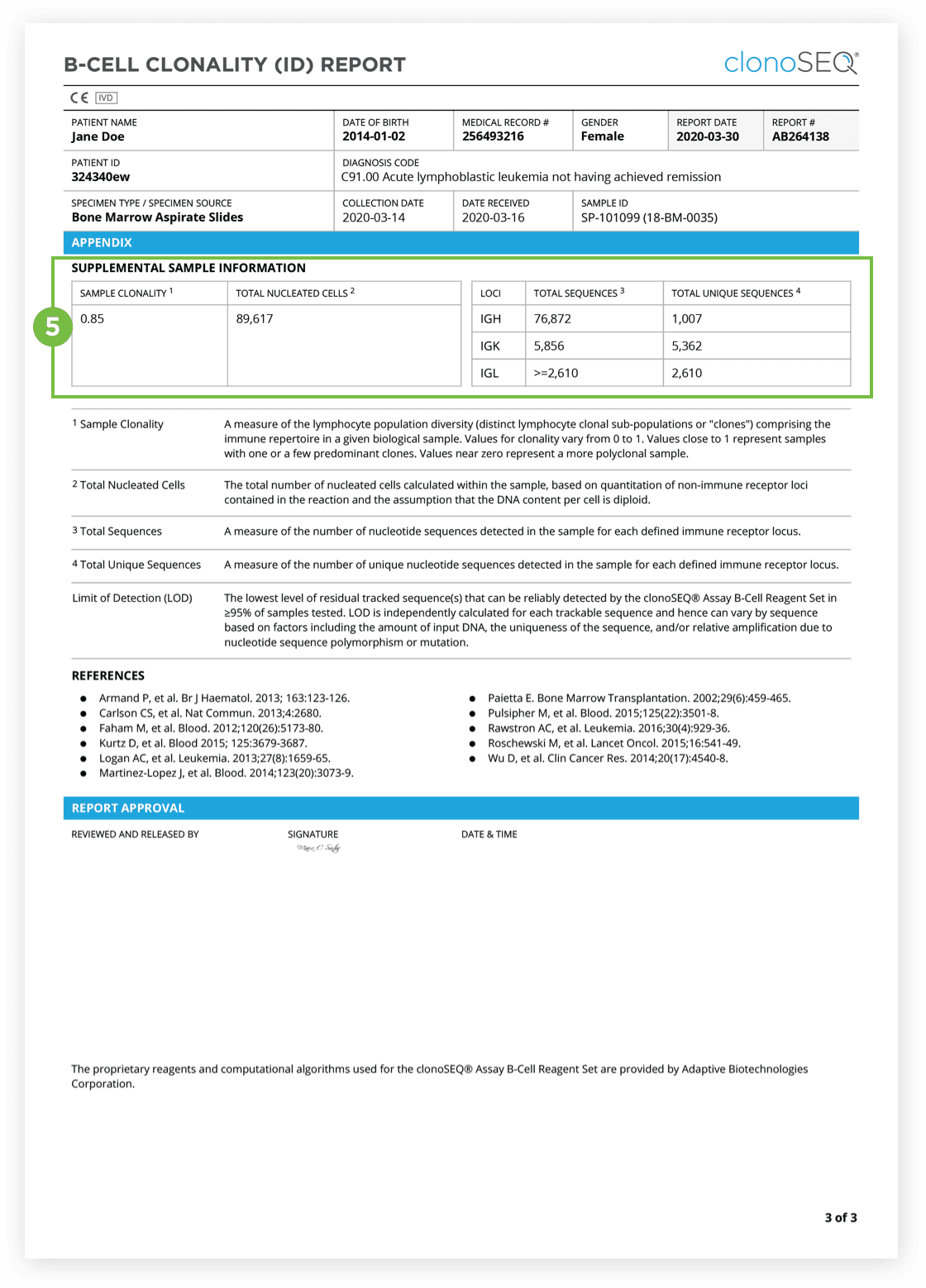
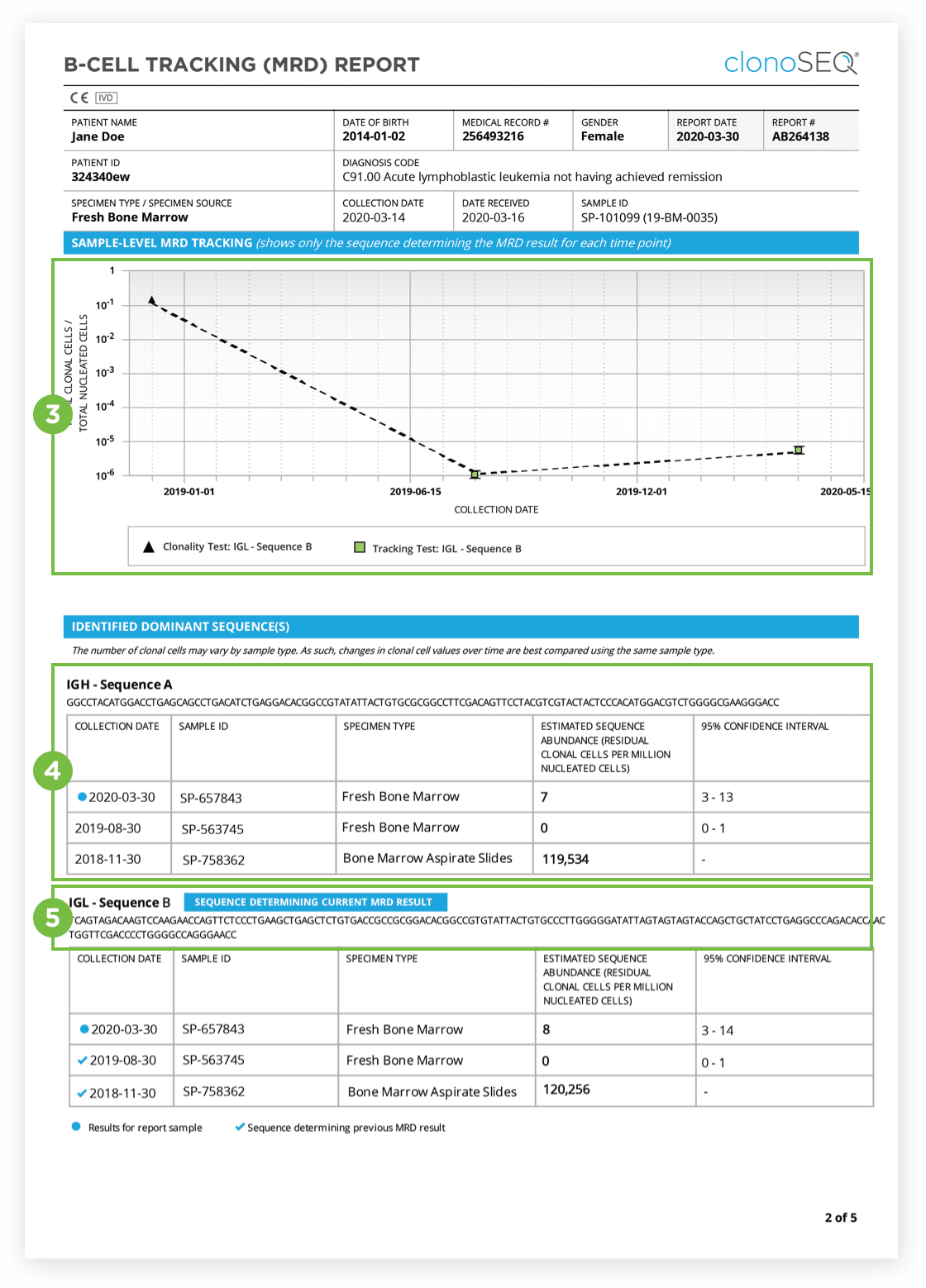
The Clonality (ID) Test is used to identify dominant DNA sequence(s) in a high disease load diagnostic sample.
Identification of at least one dominant DNA sequence is a prerequisite to future monitoring of MRD. After the dominant DNA sequences has been identified utilizing the Clonality (ID) Test, subsequent monitoring of the associated clone(s) can be completed by ordering Tracking (MRD) Tests.
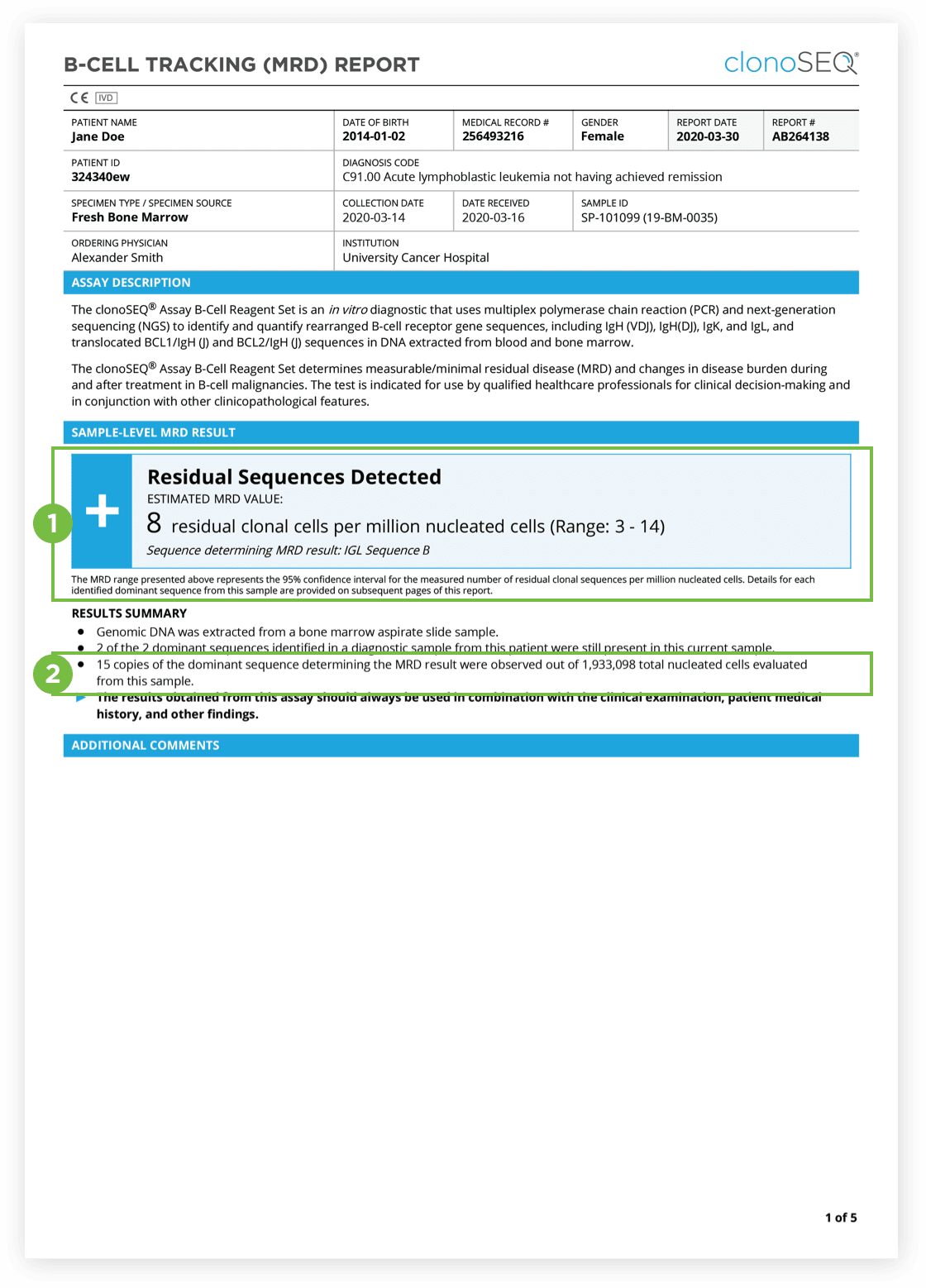
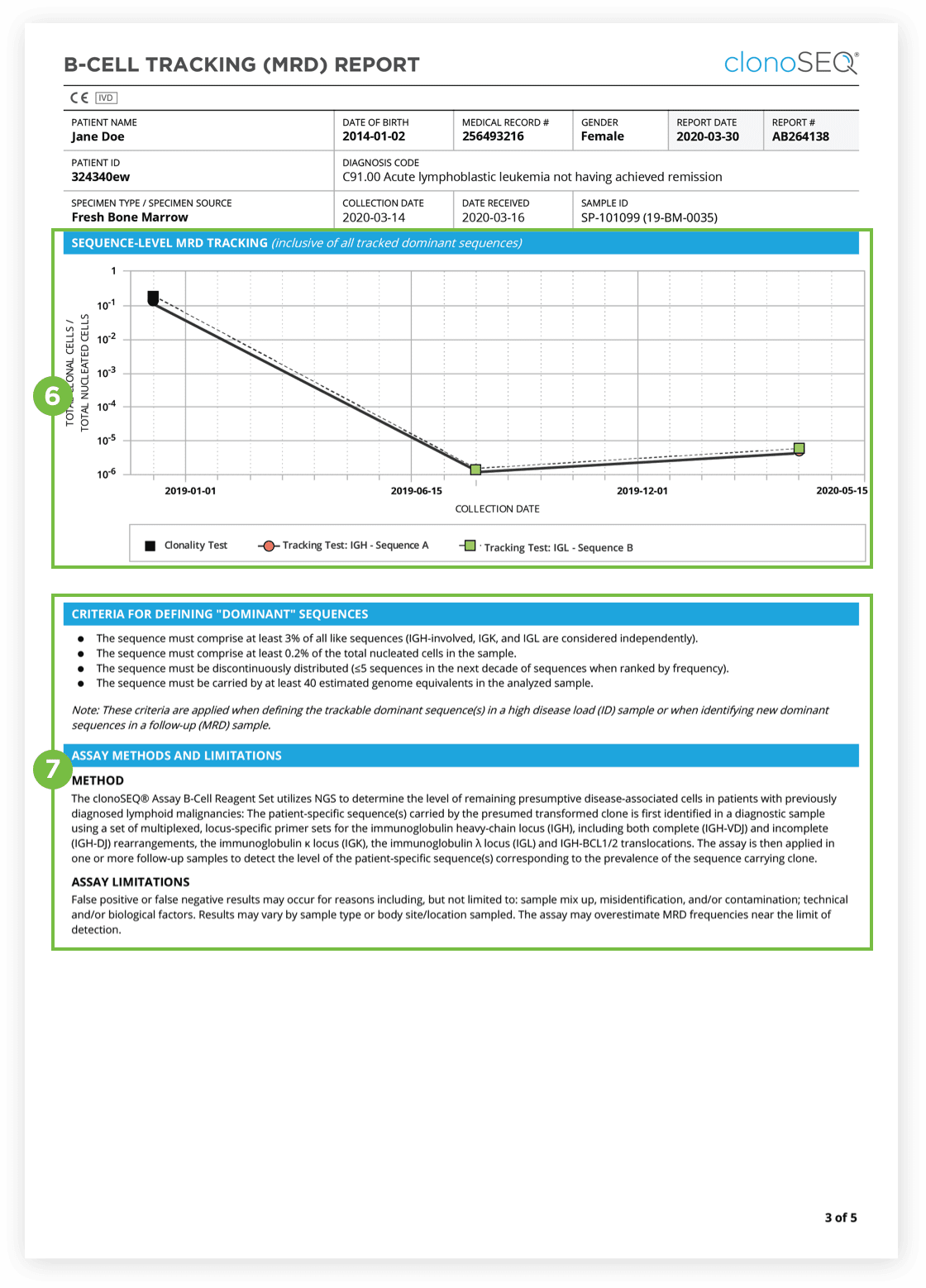
The clonoSEQ B-cell Tracking (MRD) Report provides results based on analysis of the IgH, IgK and IgL loci as well as Bcl1 and Bcl2 translocations.
After the dominant DNA sequence(s) has been identified utilizing the Clonality (ID) Test, subsequent monitoring of the associated clone(s) can be completed by ordering Tracking (MRD) Tests throughout treatment.
Explore clonoSEQ clinical reports in more depth
This page is intended for use by healthcare professionals outside of the United States.
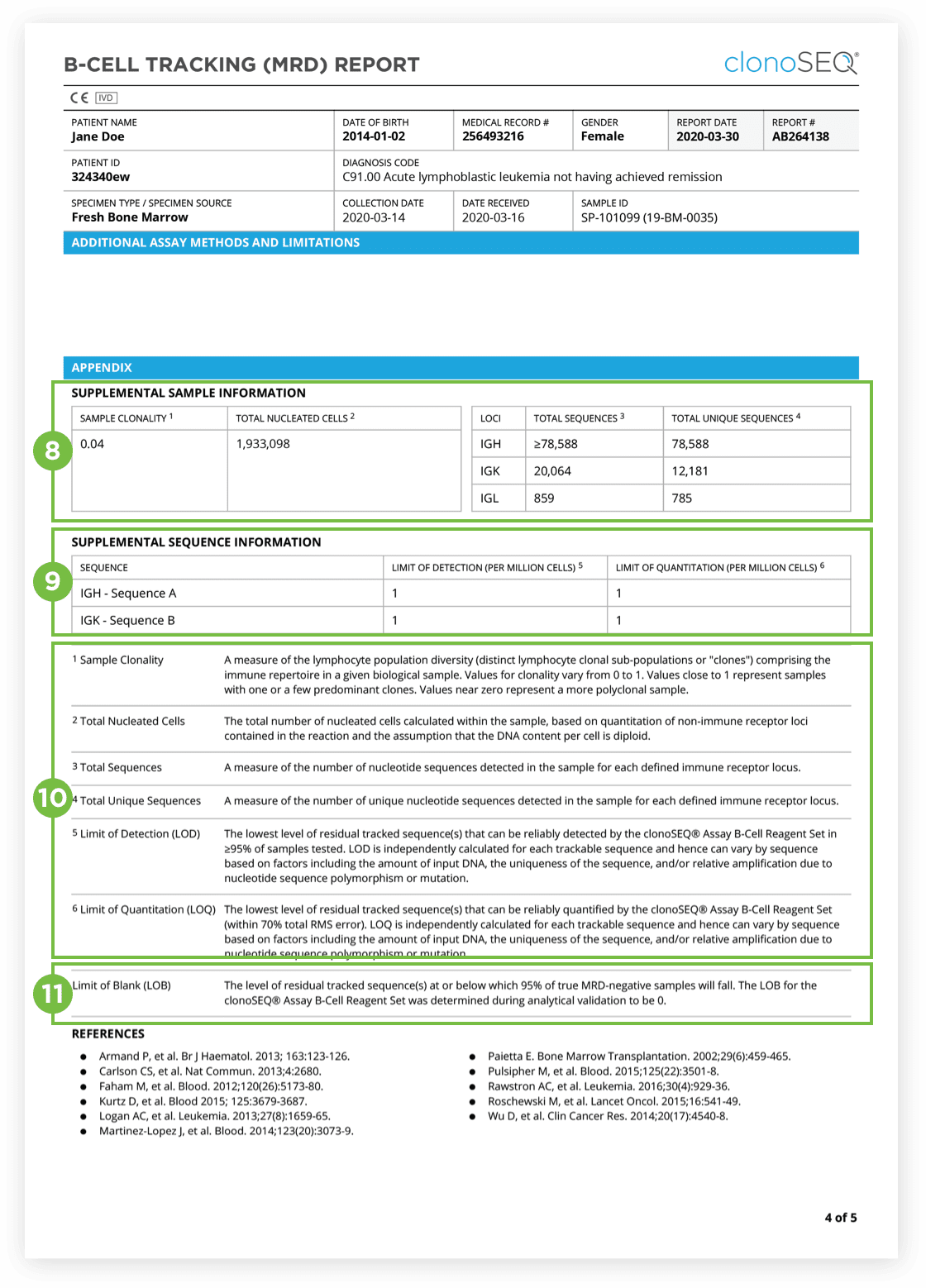
The clonoSEQ Assay B-cell Reagent Set is an in vitro diagnostic that identifies and quantifies rearranged B-cell receptor gene sequences in DNA extracted from blood and bone marrow.
It is a manual test that determines measurable/minimal residual disease (MRD) and monitors changes in disease burden during and after treatment in B-cell malignancies. The test is indicated for use by qualified healthcare professionals for clinical decision-making and in conjunction with other clinicopathological features.
The clonoSEQ Assay is being utilized for a variety of investigator-sponsored clinical trials in B-cell lymphoid cancers. If you are interested in learning more about use of clonoSEQ in your own trials, contact dxsupport@adaptivebiotech.com.
Contact Us
Make an inquiry to learn more about our products.
Citations
- The clonoSEQ Assay B-Cell Reagent Set is CE marked as in vitro diagnostic (IVD) for assessing the MRD status and changes in disease burden during and after treatment in B-Cell malignancies in DNA extracted from blood and/or bone marrow samples.